mod1 <- glm(TenYearCHD ~ age + sex + BPMeds + diabetes, data=Framingham,
family=binomial)Lesson 35: Worksheet
THIS WORKSHEET STILL IN DRAFT FORM. SKIP IT in favor of the in-class activity.
Build a model to stratify risk of CHD
Simple threshold choice
Scores <- model_eval(mod1, interval="none")
ggplot(Scores, aes(x=as.factor(.response), y=.output)) + geom_jitter(alpha=0.1, width=0.2) +
geom_violin(fill="blue", alpha=0.2)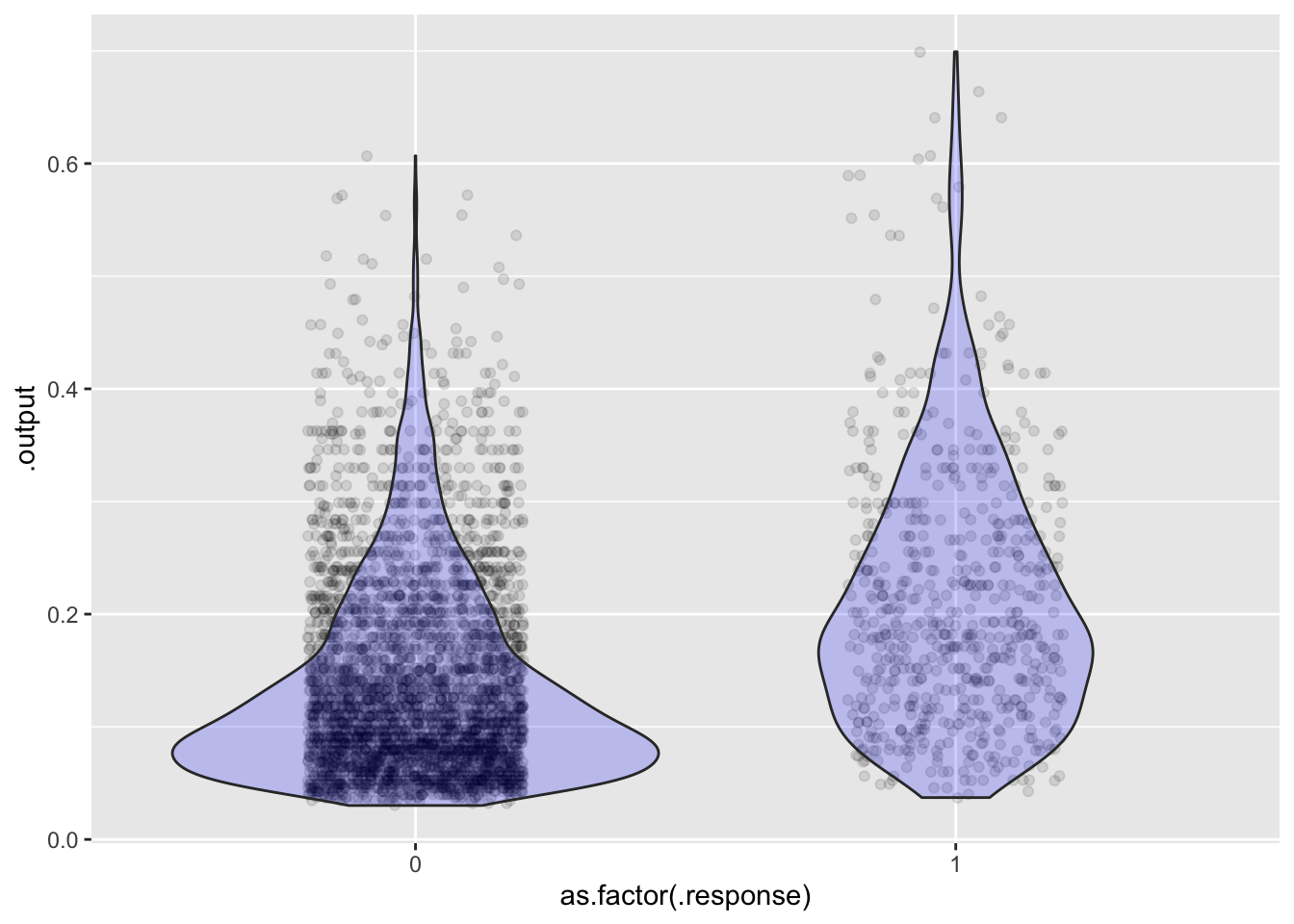
Pick a threshold
Let’s use 0.2
Construct the confusion matrix
And the sensitivity and specificity
Scores |> mutate(result=ifelse(.output > 0.2, "P", "N")) |>
group_by(.response, result) |>
tally() |>
group_by(.response) |>
mutate(frac = n/sum(n))# A tibble: 4 × 4
# Groups: .response [2]
.response result n frac
<dbl> <chr> <int> <dbl>
1 0 N 2767 0.779
2 0 P 785 0.221
3 1 N 342 0.540
4 1 P 291 0.460By changing the threshold, you can improve sensitivity at the cost of specificity and vice versa.
This isn’t a very good classifier, but let’s go with it.
Prevalence
In these training data, the prevalence of CHD is 15%.
Framingham |> summarize(prev = mean(TenYearCHD))# A tibble: 1 × 1
prev
<dbl>
1 0.152But let’s assume that the actual prevalence is 2%. Calculate the population false-positive and false-negative rates.